II. Definitions
- Pyrimidine
- Single ringed hexagonal structure forming base components of DNA and RNA (in addition to Purines)
- DNA is composed of the Pyrimidine based cytosine and thymine, and while RNA is composed of cytosine and uracil
- Purine
- Two ringed structures (Hexagon and Pentagon) forming base components of DNA and RNA (in addition to Pyrimidines)
- DNA and RNA are composed of the Purine based Adenine and Guanine
- Nucleoside
- Glycosylamine consisting of a base (Pyrimidine or Purine) combined with a sugar (ribose or deoxyribose)
- Ribose is the sugar moiety in RNA and 2-deoxyribose in DNA
- Nucleotide
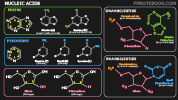
- Nucleotides combine a Nucleoside with a phosphate moiety
- DNA and RNA are composed of Nucleotides strung together
- DNA is composed of the Pyrimidine Nucleotides cytosine and thymine and purine Nucleotides Adenine and Guanine
- RNA is composed of the Pyrimidine Nucleotides cytosine and uracil and purine Nucleotides Adenine and Guanine
- Nucleic Acid
- Linear sequence of Nucleotides to form either DNA or RNA
- Base Pairs
- Pyrimidine Nucleotides Thymine (T, in DNA) and Uracil (U, in RNA) pair with the purine Nucleotide Adenine (A)
- Pyrimidine Nucleotide Cytosine (C) pairs with the purine Nucleotide Guanine (G)
III. Physiology: DNA and RNA
- DNA Replication in Cell Division
- Prior to cell division, a complete copy of the genome in DNA must be created
- Pre-initiation complex forms
- DNA double helix is unwound into two, complementary single strands by the enzyme helicase
- Helicase creates a replication fork on the DNA strand, that unravels in both directions
- Topoisomerase (DNA gyrase) facilitates straight, less coiled DNA single strands
- Primase synthesizes short primer sequences of RNA on the DNA single strand
- Primer sequences mark starting sites for DNA polymerization
- DNA double helix is unwound into two, complementary single strands by the enzyme helicase
- Elongation
- DNA Polymerase
- Starts at a primer site, facilitating polymerization of each Nucleotide sequentially, forming a new DNA fragment
- Mismatched Nucleotides are hydrolyzed and removed by DNA Polymerase
- DNA Ligase
- Connects the DNA fragments ("Okazaki Fragments") synthesized by DNA Polymerase into a single DNA strand
- DNA Polymerase
- DNA Cleanup
- Endonucleases
- Hydrolyze Nucleotide connections at the start of a DNA fragment to be excised
- Allows for excision of defective DNA segments, both during replication, as well as general DNA housekeeping
- Exonucleases
- Hydolyze terminal connections of a DNA fragment to be excised
- Endonucleases
- Resources
- DNA Replication
- DNA Transcription to RNA
- Initiation
- Core Promoters are located near transcription starting points
- General Transcription Factors and enhancers trigger the start of transcription
- RNA Polymerase
- Begins transcribing the DNA at a promoter site, and reads until it reaches a stop sequence
- Facilitates polymerization of each Nucleotide sequentially, forming a new RNA fragment
- RNA Types
- Messenger RNA (mRNA)
- Primary template for Protein synthesis, to be translated by ribosomes
- Transfer RNA (tRNA)
- Three Nucleotide sequences (Anticodons) are each matched to a specific Amino Acid (see below)
- tRNA is assembled (with an Anticodon attached to an Amino Acid) with the enzyme Amino Acyl-tRNA synthetase
- Anticodons match the 3 Nucleotide codons on mRNA
- During translation at the ribosomes, tRNA attaches to corresponding mRNA, creating an Amino Acid chain (Protein)
- Ribosomal RNA
- Forms an important component of the ribosome
- Includes ribozymes (peptide bond catalysts)
- Messenger RNA (mRNA)
- Resources
- DNA Transcription
- Initiation
- RNA Translation to Protein
- mRNA is translated by Ribosomes into Protein
- Each codon (3 Nucleotide sequence) within mRNA is matched to an anti-codon of tRNA linked to an Amino Acid
- Twenty Amino Acids are each assigned at least 2 to 3 matching codons (61 codons out of 64 possible sequences)
- The 3 remaining codons (of 64 total) function as stop signals
- tRNA assembly (Anticodon with attached Amino Acid) is synthesized via enzyme Amino Acyl-tRNA synthetase (see above)
- Resources
- RNA Translation
IV. Physiology: Nucleotides
-

- Purines (Adenine and Guanine)
- Purine synthesis via two pathways
- Salvage of already formed Purines combined with Phosphoribosyl Pyrophosphate (PRPP)
- Multistep synthesis from Glycine, Tetrahydrofolate, Glutamine, CO2, Aspartate
- Purine metabolism
- Urate formation, that is renally excreted
- Purine synthesis via two pathways
- Pyrimidines (Cytosine, Thymine, Uracil)
- Synthesis of other molecules from Nucleotides
V. References
- Goldberg (2001) Biochemistry, Medmaster, Miami, p. 36-9
