II. Pathophysiology: General
- HIV is a single-stranded RNA Retrovirus, a Sexually Transmitted Infection as well as bloodbourne pathogen
- HIV 1 is the most common worldwide HIV form and is the major cause of AIDS
- HIV 2 causes a similar presentation to HIV 1 and is found in South Africa and India
- HIV is a Retrovirus (Retroviridae)
- Retroviruses transcribe DNA from single stranded RNA (+ssRNA) via their own reverse transcriptase enzyme
- Virus then inserts its DNA into the host DNA via integrase enzyme
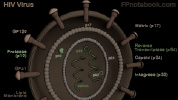
- HIV Structure
- HIV Is a spherical enveloped virion with a central cylindrical nucleocapsid
- Outer wall of HIV is a lipid bilayer membrane
- Membrane contains embedded Glycoproteins (gp120 is external, gp41 is transmembrane)
- P10 protease is present between this outer wall and the nucleus-like structure's capsid membrane
- Nucleus-like structure is surrounded by capsid wall (p17) and contains RNA and enzymes
- RNA Dimer: 2 identical strands of single stranded RNA
- Three Enzymes: p32 Integrase, p64 reverse transcriptase, protease
- Nucleocapsid composed of capsid Proteins (CA, esp. p24) surrounds the RNA dimer and enzymes
- Images
III. Pathophysiology: HIV Infection
- HIV infects T Helper Cells (CD4+ Cells)
- Host Cell Binding
- HIV gp160 (gp120 + gp41) binds to CD4 receptors on T Helper Cells
- Gp160 also binds tp Macrophages, Monocytes and CNS Dendritic Cells
- Host Cell fusion and penetration
- Virus envelope fuses with the host cell membrane, and RNA infiltrates the host cytoplasm
- Fusion Inhibitors (e.g. Enfuvirtide) blocks HIV fusion to cell anchor Proteins
- CD4 Coreceptors (CCR5 or CXCR4) must be present on the host cell surface for HIV penetration
- Decreased HIV risk in patients with lower levels of CCR5 on CD4+ cells
- Lymphocyte derived Proteins (e.g. RANTES, MIP1) bind CCR5 and also reduce HIV risk
- CCR5 Inhibitors (Maraviroc) block HIV binding at CCR5 co-receptor
- Virus envelope fuses with the host cell membrane, and RNA infiltrates the host cytoplasm
- Proviral DNA generation
- Complementary DNA (cDNA) is formed from HIV RNA via reverse transcriptase enzyme
- cDNA penetrates the host cell's nucleus and integrates with host DNA via HIV integrase enzyme
- Blocked by Integrase Inhibitors (e.g. Raltegravir, Dolutegravir)
- Integrated DNA may remain dormant until activated for viral replication
- Activation may be triggered by T Cell activation by coinfection (e.g. Tb, CMV, PJP, HSV)
- T Cell activation releases Proteins that bind HIV LTR and lead to HIV DNA Transcription
- HIV DNA Transcription and translation
- Proviral DNA is transcribed into Messenger RNA (mRNA) by host RNA Polymerase
- mRNA is translated into long chains of HIV viral Proteins
- Long Protein chains are then cleaved by proteases into shorter HIV Proteins
- Blocked by Protease Inhibitors (e.g. Ritonavir, Indinavir, Lopinavir)
- HIV Budding
- Single stranded RNA and viral Protein enzymes are repackaged into virions
- Virions bud through the host cell surface, taking with them part of the host cell membrane
- HIV specific Glycoproteins are embedded in the outer membrane
- T cells die as thousands of virions are released
- T Cell Death Mechanisms
- HIV Virion budding (see above)
- Multinucleated Giant cells
- Infected T Cells may fuse with other T Cells via surface binding to gp160
- Fused cells form multinucleated giant cells (syncytial giant cells)
- HIV may be passed from cell to cell via fusion, bypassing antibodies external to the cells
- CD8+ Cytotoxic T Cell response
- Cytotoxic T cells may destroy infected CD4+ Cells marked by surface gp160
- Bystander CD4+ Helper T Cell Death
- Other immune cell HIV effects
- B Cell Dysfunction
- Polyclonal B Cell activation by HIV results in hypergammaglobulinemia
- Diminished response to new Antigens (e.g. infections, Immunizations)
- Autoantibody formation resulting in Autoimmune Conditions
- Monocyte and Macrophages as trojan horse, HIV reservoirs
- HIV actively replicates with these cells without host cell destruction
- Monocyte and Macrophages may cross the blood brain barrier and expose the CNS to HIV
- B Cell Dysfunction
- Reactivation and Propagation
- HIV proviral DNA is activated after a latent period of months to years
- Reactivation and propagation via HIV Budding results in progressive CD4+ T Cell destruction
IV. Pathophysiology: HIV Genome
-
Retrovirus Genome Sequences (common to all Retroviruses)
- Long Term Repeat Sequences (LTR)
- LTRs are found at each end of the transcribed DNA strands, flanking the intervening genes
- LTRs serve 2 purposes
- Sticky Ends
- Serves as a target for the integrase enzyme
- Allow for easier insertion into host DNA
- Promotor
- Enhances viral DNA Transcription after it has been inserted into host DNA
- Sticky Ends
- Group Antigen (gag) codes for major Retroviral structural Proteins
- DNA Polymerase (pol) codes for major Retroviral enzymes
- P64 reverse transcriptase
- P32 Integrase
- Protease
- Protease cleaves gag and pol Proteins from the precursor molecules (enzyme activation)
- Envelope Protein (env) codes for surface Glycoproteins
- GP160 binds CD4 receptors
- GP 160 is composed of 2 subcomponents
- GP120 (head)
- GP41 (stem)
- Long Term Repeat Sequences (LTR)
- Early HIV Specific Genome Sequences
- Transactivator Protein (tat)
- Binds viral genome and activates transcription
- Regulator of Expression of Virion Proteins (rev)
- Binds the rev response element (RRE) within the Env gene
- Increases reading of gag, pol and env, increasing generation of HIV virions
- Negative Factor (nef)
- Decreases CD4 and MHC1 expression on surface of infected CD4+ Cells
- Suppresses cytotoxic T cell response (CD8) that would otherwise kill HIV infected cells
- Transactivator Protein (tat)
- Late HIV Specific Genome Sequences
- Virion Infectivity Factor (vif)
- Integral to dsDNA generation from HIV RNA
- Blocks APOBEC3
- APBEC3 is an innate viral defense, restricting viral replication
- APBEC3 modifies DNA, changing cytosine to uracil (resulting in G-A mutation)
- Viral Protein R (vpr)
- Regulates nuclear import of HIV1
- Allows for HIV replication within non-dividing cells (e.g. Macrophages)
- Viral Protein U (vpu)
- Decreases CD4 and MHC1 expression on surface of infected CD4+ Cells
- Enhances HIV virion release from infected cells
- Virion Infectivity Factor (vif)
V. Pathophysiology: HIV Genome Heterogeneity
- HIV replication is subject to frequent mutations and significant genetic variation (heterogeneity)
- Several HIV genes have hypervariable regions with high mutation rates
- Env gene mutations (esp. gp120 encoded region)
- Reverse Transcriptase mutations
- Major HIV subgroups based on gag and env Protein variation have been developed (Groups A-K)
- Subtype B is most common in North America and Europe
VI. Course
- See HIV Course
VII. References
- Gladwin, Trattler and Mahan (2014) Clinical Microbiology, Medmaster, Fl, p. 268-75
- Mahmoudi (2014) Immunology Made Ridiculously Simple, MedMaster, Miami, FL
- (2016) Transfus Med Hemother 43(3):203-22 +PMID: 27403093 [PubMed]
- McLaren (2021) Nat Rev Genet 22(10):645-57 +PMID: 34168330 [PubMed]